FAQs - Frequently Asked Questions
1) How do I create a STUDY set?
You can create a study set using the EEGLAB GUI or the function pop_createstudy(). There is an extra function in the CORRMAP plugin distribution called create_study_fv() that allows you to easily create a STUDY set using a group of datasets stored in a specific folder. To learn how to use this function check >> help create_study_fv.
2) How do I pre-cluster a STUDY? Do I need to pre-cluster the STUDY before running CORRMAP?
To pre-cluster your STUDY set please use the STUDY menu in EEGLAB GUI or the STUDY functions (check >> help std_preclust or >> help pop_preclust). We recommend that when you pre-cluster you choose at least these 3 parameters: ERPs, spectra, topo. The pre-clustering step is not mandatory to run CORRMAP. It is only necessary if you wish to save ICs found by CORRMAP in a cluster in the STUDY structure. When you do this you can use the STUDY plot functions to edit/explore the cluster. See an example here.
3) How do I choose a template?
Before using CORRMAP you should have inspected the ICs obtained after running ICA for at least one dataset. We recommend that you choose templates representative of biological artifacts such as eye blinks, lateral eye movements or heartbeat artifacts. There are many plotting options that allow you to check if an IC represents a particular artifact. If you are not familiar with ICA and inpection of ICs, please check the EEGLAB tutorial.
4) How do I choose a correlation threshold?
If you are not an experienced ICA user, the best option is to run CORRMAP in automatic mode but it is important to regard the automatic threshold as a first guiding value only. In some situations it may be necessary to adjust the threshold after inspection of the cluster initially obtained. The output plots provide valuable information to help you to adjust the threshold. For more information check >> help corrmap or the reference paper [1].
5) Which number of ICs shall I choose?
Current CORRMAP version can select up to three ICs with the largest supra threshold correlation with the template. We recommend using the default value: 2 ICs per dataset. However you can easily change this parameter and adapt it to suit your data. For more details check >> help corrmap or the reference paper [1].
6) Why do I need to create the field EEG.badchannels for the datasets where I have removed channels?
The indices of channels removed from datasets must be saved in the EEG structure in a field called EEG.badchannels. This step must be done before creating your STUDY set. These indices are used as a parameter for the function interp_chan(). This function interpolates the ICA inverse weigths so that CORRMAP output is shown correctly. For more details check >> help corrmap or >> help interp_chan.
7) Why shall I store EEG.badcomps?
When you store the indices of bad components, the EEGLAB feature Tools > Remove Components for each single dataset can then be used to remove these components from the dataset (see below).
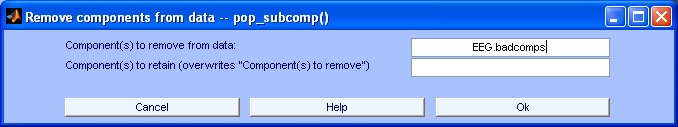
If you haven’t selected bad components a priori, you need to create an empty field (EEG.badcomps=[];) in the EEG structure of each single dataset present in your STUDY. If you don’t like scripting you can use the same function for simultaneously creating your STUDY structure and the empty field EEG.badcomps for all datasets. (check >> help create_study_fv).
Disclaimer: The old datasets are overwritten so it is a good idea to keep a backup copy in a separate folder. It is only possible remove EEG.badcomps when processing single datasets. This option is not available when processing the STUDY set. It must be performed afterwards individually for each dataset. It might happen that duplications of the ICs indices are stored. If that occurs you can still use the "Remove Components" option, since it ignores duplications.
8) Can I run CORRMAP from the command line?
Yes, you can. To learn how to do that check >> help corrmap. When running CORRMAP from the command line you should bare in mind that the parameter for the correlation threshold (‘th’) must be a string. See example below.
>> CORRMAP=corrmap(STUDY,ALLEEG,1,5,’th’,’0.85’,’ics’,1);
When running CORRMAP from the command line it is also possible to change the plot option to ‘none’, so the output plots are not displayed.
>> CORRMAP=corrmap(STUDY,ALLEEG,1,5,’pl’,’none’);
To learn about CORRMAP ouputs check FAQ number 9.
9) What are the outputs of CORRMAP?
The main ouput is a summary plot showing the template, the selected ICs, their correlations with the template and further cluster information. If you want to save the output plot, you need to use the standard MATLAB commands from the menu or from the command line. For more information refer to MATLAB help documentation.
When CORRMAP is run from the command line, an output structure with the name CORRMAP is saved in MATLAB working space. This structure contains six main fields where information about input parameters and output parameters is stored. You can find more about the CORRMAP structure fields here.
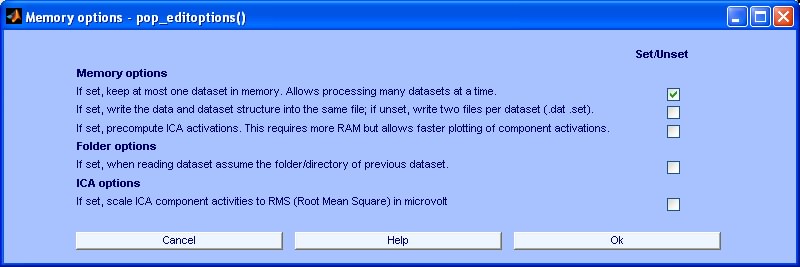
10) How do I deal with "Out of Memory" error messages?
CORRMAP is a tool entirely based on the STUDY structure, so please make sure that you choose the right memory options (File > Memory and other options) when using it. We recommend that you use the following options:
11) I can't find an answer to my problem in this list. What shal I do?
If you have checked all CORRMAP documentation available (tutorial, FAQs and help messages) and you still haven't found a solution to your problem, please send an email to filipa dot viola at uni-oldenburg dot de. However please be aware that it might take some time till we are able to answer.
This page was last updated May 20, 2011.